Introduction
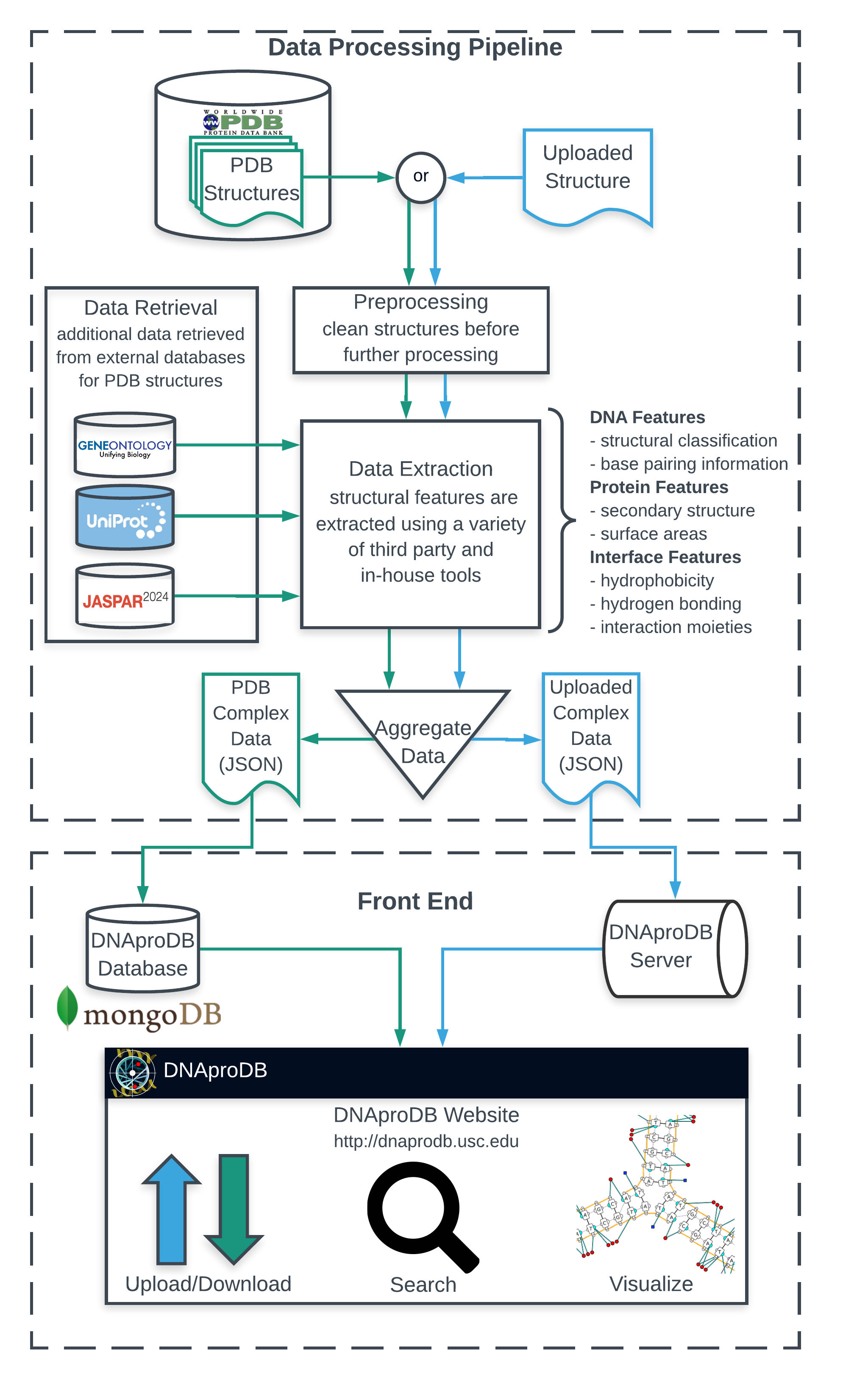
DNAproDB is processing pipeline, web server and database which aims to assist researchers in performing structural analysis of DNA-protein complexes. The DNAproDB processing pipeline takes as input the three-dimensional structure of a DNA-protein complex (in mmCIF or PDB format) and extracts useful bio-physical information from it, using a combination of our own tools and well known third-party libraries and software. Information is organized into three main categories: DNA features which describe the sequence and structure of the DNA present in the complex, protein features which describe the sequence and structure of the protein in the complex, and interface features that describe the various DNA-protein interactions and statistics of those interactions present in the complex. This information is then combined in a hierarchical way and stored in a JSON file, which is ideal for easy parsing and integration with the web.
DNAproDB provides tools for exploring and visualizing the data extracted from a complex. These tools are found in the report page for each structure, which can be accessed by searching the database, typing the PDB identifier in the search box at the top of any page on the website, or by following the link generated when uploading a structure. These tools allow you to visualize various DNA-protein interactions side-by-side with the three-dimensional view of the complex. Clicking and hovering on different parts of the visualizations will display additional information or highlight parts of the three-dimensional structure and allow the user to visually explore different layers of the DNA-protein interface. These visualizations can be customized and exported as a PNG file.
DNAproDB is also a database. We have pre-processed a large number of DNA-protein complexes retrieved from the Protein Data Bank (PDB) which we store in a database - hence the "DB" in "DNAproDB". The database is designed as a document-oriented database, and is implemented in MongoDB. The data stored in the DNAproDB database is identical to the data produced by the processing pipeline and the data available in the report pages, and is returned in the form of a JSON file.
On the remainder of this page you'll find information about how to use the various aspects of DNAproDB. Have a scroll through or use the side-navigation to the left to jump to a specific section.
Using Data from DNAproDB
Any page which displays information to the user about a DNA-protein complex is generated from the data stored in a DNAproDB data file (JSON format) corresponding to that complex. The data files are self-contained, that is, a single data file describes all the information about a particular DNA-protein complex structure in one place. For structures which come from the PDB, this includes a small amount of information which is retrieved from external databases such as UniProt, CATH and the Gene Ontology knowledgebase. This information is not included for uploaded structures since it is assumed these are unpublished - if they are, search the database for them instead. Users can download our data for their own purposes offline (all we ask is a citation). You can download the entire database as a flat file from the download page, download data for a set of structures returned from a query by searching the database, or can visit the report page for an individual structure and download the data there.
In order to understand the way DNAproDB data is organized, it is important to define some key terminology which is used throughout the site and to describe the conceptual framework DNAproDB is built on. Below is a diagram representing the conceptual framework of how we organize structural data in DNAproDB, which is based on the Protein Data Bank's framework.
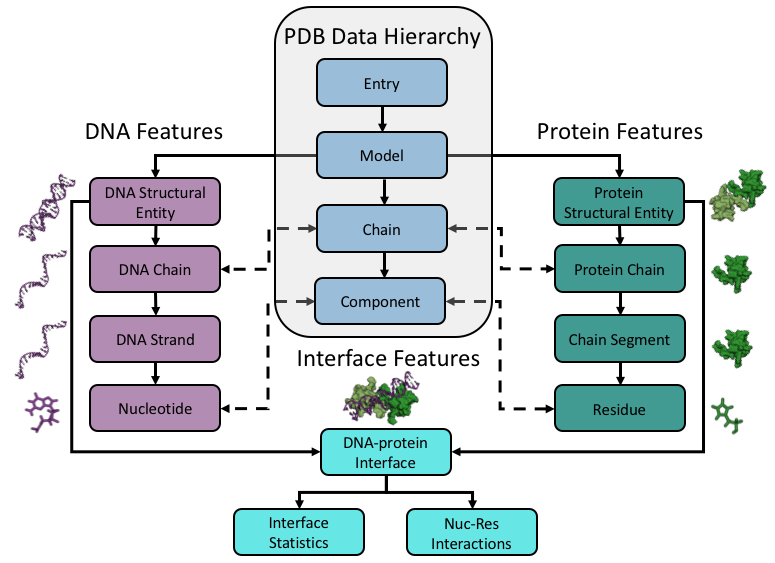
Below is the basic schema for the way we organize the data in DBAproDB. In general, every DNA-protein complex which is stored in DNAproDB is represented by a JSON file with the same basic structure. However, some fields are of variable length, which depends on the size and complexity of the structure. Use the interactive diagram below to explore the structure of the data file.
As a more concrete example, below is a JSON file which is returned from DNAproDB for the small structure 1jgg. Click on the different fields to expand them and explore the data structure.
We use various abbreviations in feature names throughout the DNAproDB data files. Reference the table below for the meaning of these abbreviations.
| abbreviation | full name | description |
|---|---|---|
| nuc | nucleotide | A DNA nucleotide (nucleic acid). |
| res | residue | A protein residue (amino acid). |
| pp | phosphate | A DNA backbone structural moiety which consists of the phosphate group. |
| sr | sugar | A DNA backbone structural moiety which consists of the pentose sugar group. |
| wg | major groove or "wide groove" | A helical DNA base structural moiety which consists of the base edge that is exposed in the major groove. |
| sg | minor groove or "small groove" | A helical DNA base structural moiety which consists of the base edge that is exposed in the minor groove. |
| bs | base | A DNA structural moiety which consists of the entire nucleoside base. |
| mc | main chain | A protein structural moiety which consists of the residue backbone. |
| sc | side chain | A protein structural moiety which consists of the residue side chain or functional group. |
| H | helix | Simplified protein secondary structure consisting of 'H', 'G' or 'I' helices using DSSP notation. |
| S | beta strand/beta sheet | Simplified protein secondary structure consisting of 'E' using DSSP notation. |
| L | loop | Simplified protein secondary structure consisting of 'B', 'T', 'S' and '-' using DSSP notation. Anything that is not a helix or a strand. |
| sse | secondary structure element | A continuous sequence of residues that share a helix or strand conformation. |
| basa | buried solvent accessible surface area | The amount of solvent accessible surface area which is lost when two components of the complex are bound to each other. |
| fasa | free solvent accessible surface area | The solvent accessible surface area when the component is in a "free" state (e.g. the SASA of the DNA when the protein is removed from the structure). |
| sesa | solvent excluded surface area | The solvent excluded surface area of a component. |
| hbond | hydrogen bond | A favorable electrostatic interaction between a polar hydrogen (donor) and an electronegative atom (acceptor). |
| vdw | van der waals | Two atoms which are in close proximity and may form induce dipole interactions |
| cv | circular variance | A measure of the protein surface geometry |
In addition to extracting information from a data file in-browser via the report page, one can download DNAproDB data files, which contain all the information about a DNA-protein complex provided by DNAproDB, and parse those files offline. In general, all you need is a programming language which supports a JSON parser, and the parser its self. A long list of JSON parsers is available at the official JSON web-page.
Below are some simple examples in Python. Note that neither JSON nor the structure of the DNAproDB data files are specific to python - the user can work in whatever language they prefer. When downloading data for multiple structures from the DNAproDB database, the data will be returned in a single file, which contains and array of objects. Each object corresponds to a single structure.
import json
with open("data_file.json") as DATA:
dnaprodb_data = json.load(DATA) # load the data file using the json module
hb_count = 0 # store the number of hydrogen bonds which meet our criteria
model_num = 0 # choose the first model in the structure
for interface in dnaprodb_data["interfaces"]["models"][model_num]: # iterate over each DNA-protein interface for model 'model_num'
for interaction in interface["nucleotide-residue_interactions"]: # iterate over each nucleotide-residue interaction
if(
interaction["res_name"] == "ARG" # check if the residue in the interaction is an arginine
and
interaction["nuc_secondary_structure"] == "helical" # check if the nucleotide is in a helical conformation
):
hb_count += interaction["hbond_sum"]["wg"]["sc"] # if so, add the number of major groove-side chain hbonds to count
print("Arginine - Minor Groove Hbonds: %d" % count) # print the result
import json
with open("data_file.json") as DATA:
dnaprodb_data = json.load(DATA) # load the data file using the json module
model_num = 0 # choose the first model in the structure
narrow_minor_groove_nucleotides = [] # an array to store nucleotide identifiers in narrow minor groove regions
for entity in dnaprodb_data["dna"]["models"][model_num]["entities"] # loop over DNA entities in model model_num
for helix in entity["helical_segments"]: # loop over helices for each entity
for i in xrange(helix["length"]):
if(helix["shape_parameters"]["minor_groove_curves"][i] < 5.0): # check if minor groove at position 'i' is less than 5.0
narrow_minor_groove_nucleotides.append(helix["ids1"][i]) # if so push nucleotide id at position 'i' from strand 1
narrow_minor_groove_nucleotides.append(helix["ids2"][i]) # and from strand 2
narrow_minor_groove_interactions = [] # array to store interactions meeting our criteria
for interface in dnaprodb_data["interfaces"]["models"][model_num]: # iterate over each DNA-protein interface for model model_num
for interaction in interface["nucleotide-residue_interactions"]: # iterate over each nucleotide-residue interaction
if("sg" in interaction["nucleotide_interaction_moieties"]): # check if this interaction involves the minor groove
if(interaction["nuc_id"] in narrow_minor_groove_nucleotides): # check if the nucleotide is in our list of narrow minor groove nucleotides
narrow_minor_groove_interactions.append(interaction) # if the above conditions are met, push the interaction
# now the array 'narrow_minor_groove_interactions' contains all minor groove
# interactions which involve nucleotides in narrow minor groove regions.
Searching DNAproDB
DNAproDB's database has powerful search capabilities that allow users to search for structures based on characteristics of the DNA, the protein, or the DNA-protein interface. Multiple criteria can be combined using different selection logic. In addition, you can search for structures with their PDB identifier or supply a list of PDB identifiers in combination with additional criteria, which act as a filter. This search capability is unique to DNAproDB and can be used to generate data sets or find structures based on structural and bio-physical features of the DNA, protein, or DNA-protein interactions present in the structure. The easiest way to search the database is through the search-page, where you can construct a query using a simple expandable form.
The search form can be accessed from the search page. When first presented, the form will be empty and contain no fields. Choose from one of the feature categories and you'll be presented with different fields you can fill in which correspond to the different aspects of a DNA-protein complex (protein properties, DNA properties, or DNA-protein interaction properties).
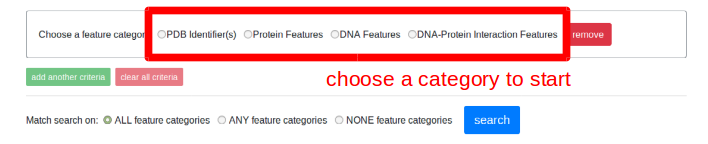
Choosing one of the feature categories will update the form with a panel which shows different fields corresponding to that category. Additionally, you may choose to add more categories, or clear all the categories with the "add another category" and "clear all categories" buttons, remove an individual category using the "remove" button, and choose how different categories are combined logically.
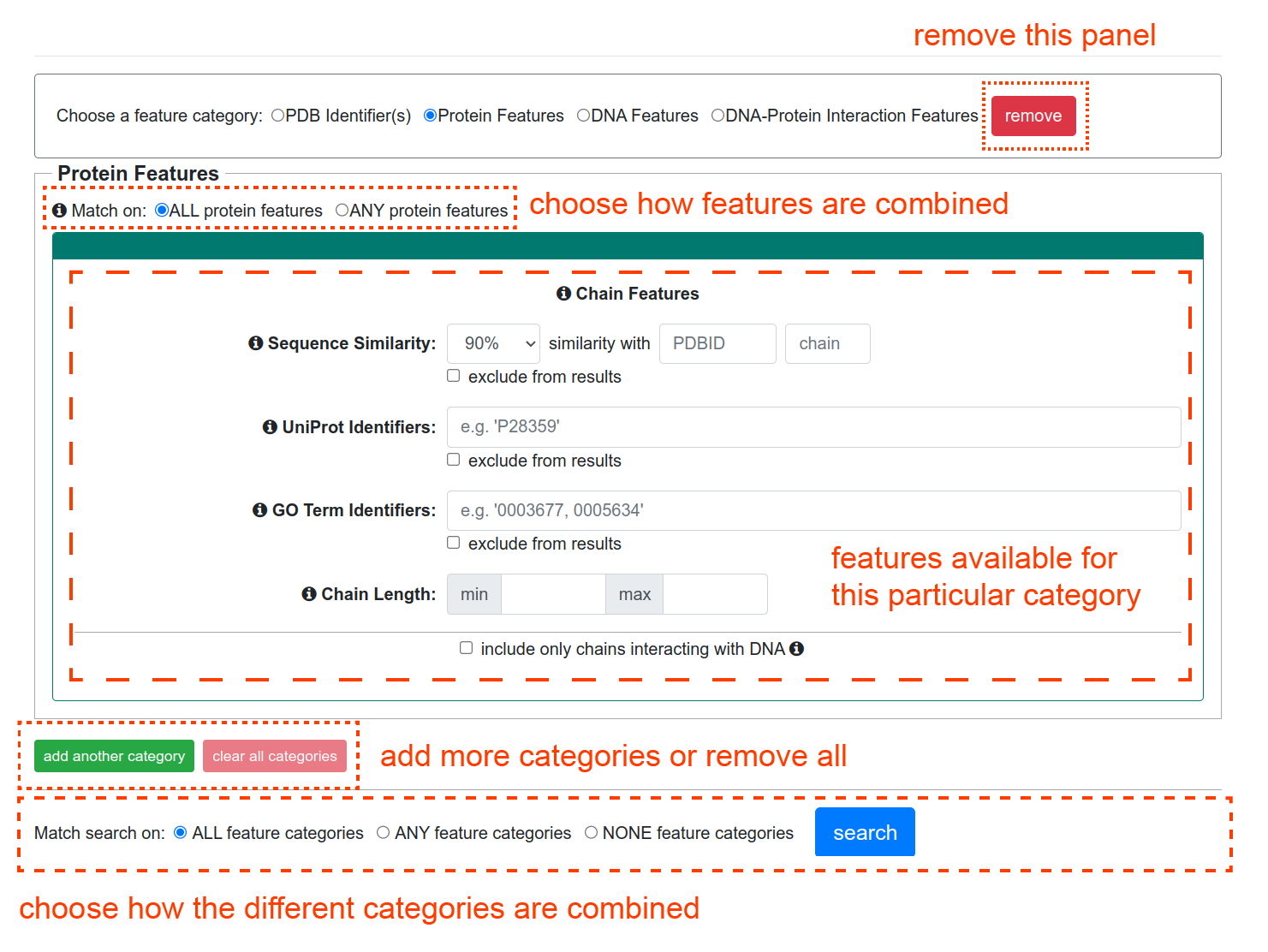
Once chosen, any fields within a category which are left blank or unchecked will be ignored, so you only have to choose items which you care about. You can choose to search for structures which match any selected feature within a category, or which match all selected features. For example, if you are matching on all selected features and choose DNA properties and specify that the sequence length should be between 10 and 20 base-pairs, and also check that the DNA Conformation should be B-form, then only structures which meet BOTH requirements will be returned. However, for fields which have multiple options such as B-DNA, A-DNA, or Other under DNA Conformation, choosing multiple options will match structures which meet ANY of the selected options. So, if you chose the sequence length to be between 10 and 20 base-pairs, and checked B-DNA and A-DNA under DNA conformation, then that will return structures in which the DNA is between 10 and 20 base-pairs in length AND is either B-DNA or A-DNA like.
The DNAproDB database can be searched using a web API, but requires one to be familiar with both the MongoDB query syntax and the DNAproDB data file structure. DNAproDB is built using MongoDB version 3.4 (documentation for their query language is available here). We provide options to search the database by passing a JSON query string to our API with an optional projection string, or to retrieve data for an individual entry by its PDB identifier. We also provide some convenience options such as getting the total number of entries in the database. All data is returned in JSON - either a single document or an array of documents. Note that the use of the $where operator in queries is forbidden - any query using this operator will be rejected. See the table below for information about using our API.
| URL |
/cgi-bin/request-data
|
|---|---|
| HTTP Method | GET | POST |
| URL Params |
Querying query=[JSON string] (required)
example: query={"protein.chains.organism": {"$regex": "homo sapiens", "$options": "i"}}
projection=[JSON string] (optional)
example: projection={"structure_id":1, "protein.chains": 1}
Retrieve a single entry pdbid_id=[four character PDB identifier]
example: pdb_id=1jgg
Get the number of entries count=[boolean]
example: count=true
Get last time the database was updated last-updated=[boolean]
example: last-updated=true
Get list of all PDB identifiers pdbid-list=[boolean]
example: pdbid-list=true
|
| Examples |
Here we use jQuery to make a POST request using the 'query' parameter to find all entries with a human protein chain. We use the $regex operator to look for protein chains where the 'organism' feature matches "homo sapiens", case-insensitive.
$.ajax({
url: "https://dnaprodb.usc.edu:/cgi-bin/request-data",
dataType: "json",
data : {
'query': '{"protein.chains.organism": {"$regex": "homo sapiens", "$options": "i"}}',
},
type : "POST",
success : function(result) {
console.log(result);
}
});
Here we send a GET request with wget to retrieve the number of entries in the database.
wget -O count.json https://dnaprodb.usc.edu/cgi-bin/request-data?count=1
cat count.json
|
| Notes |
The use of the $where operator is not allowed. Queries using this operator will be automatically rejected. When using regular expression please use the {"$regex": pattern, "$options": [options]} syntax. If you require to use regular expressions inside of an $in expression, please convert this to an a single regular expression instead. For example, rather than using {"$in": [\pattern1\, \pattern2\, exact3]}, convert this to {"$regex": "pattern1|pattern2|^exact3$"}.
|