Getting Started with DNAproDB
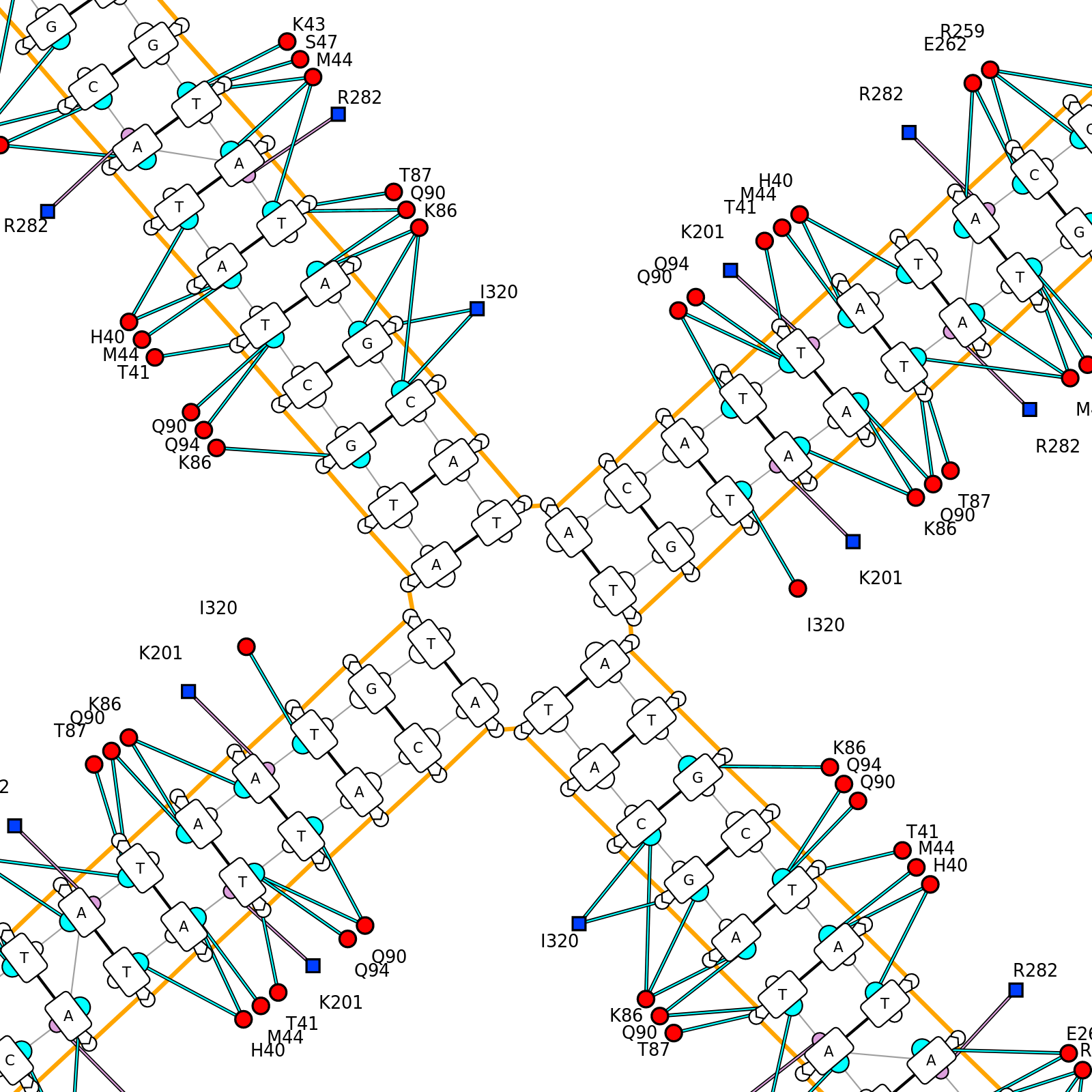
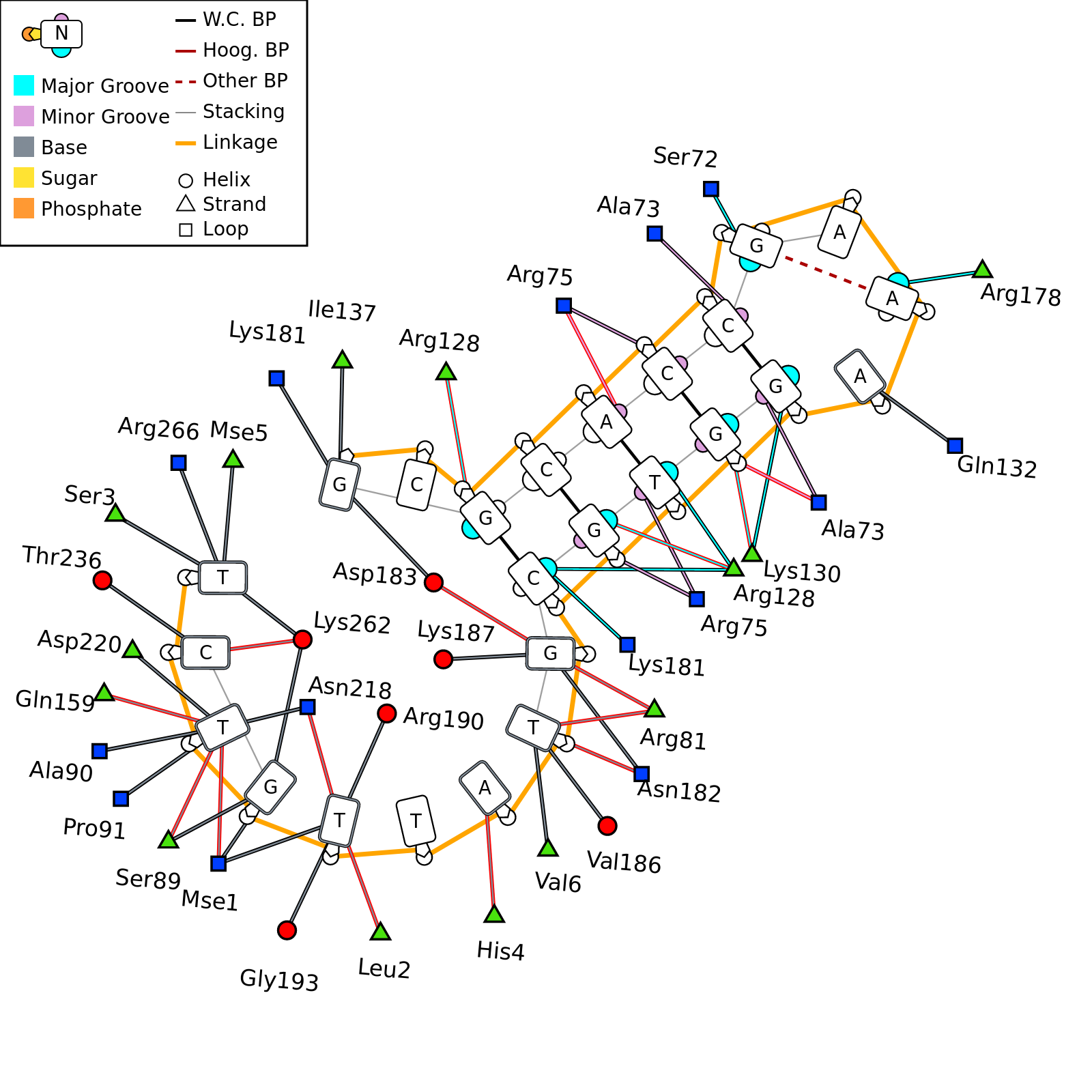
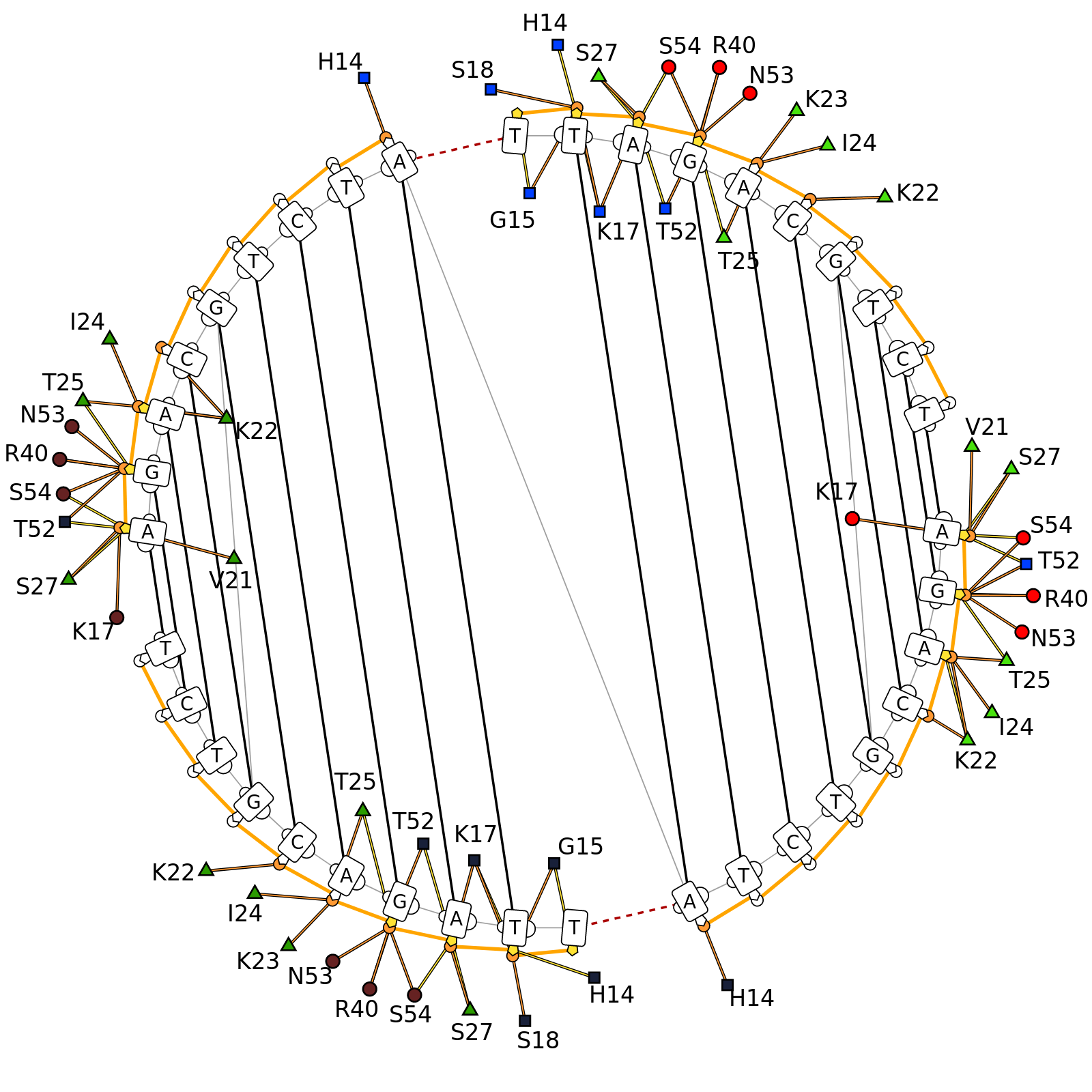
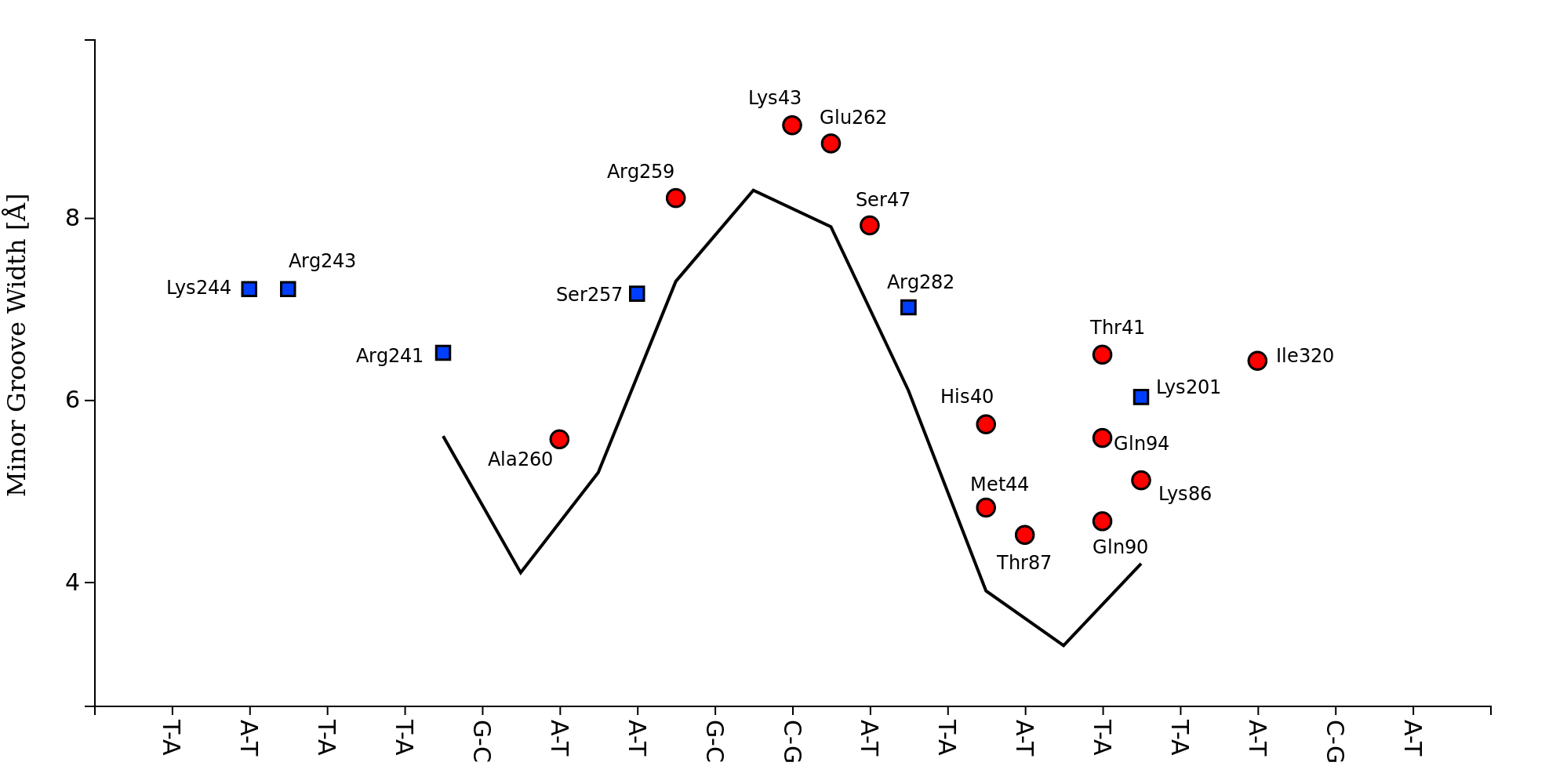
DNAproDB is a database, structure processing pipeline and web-based visualization tool which makes structural analysis of DNA-protein complexes easy. Our database contains structural and biochemical features extracted from structures of DNA-protein complexes contained in the Protein Data Bank. This data can be used to analyze individual structures or to generate large datasets by constructing queries on a set of features using the search form. Additionally, you can upload your own structure using the upload form and use the same processing and visualization tools for unpublished data.
In short, you can use DNAproDB to
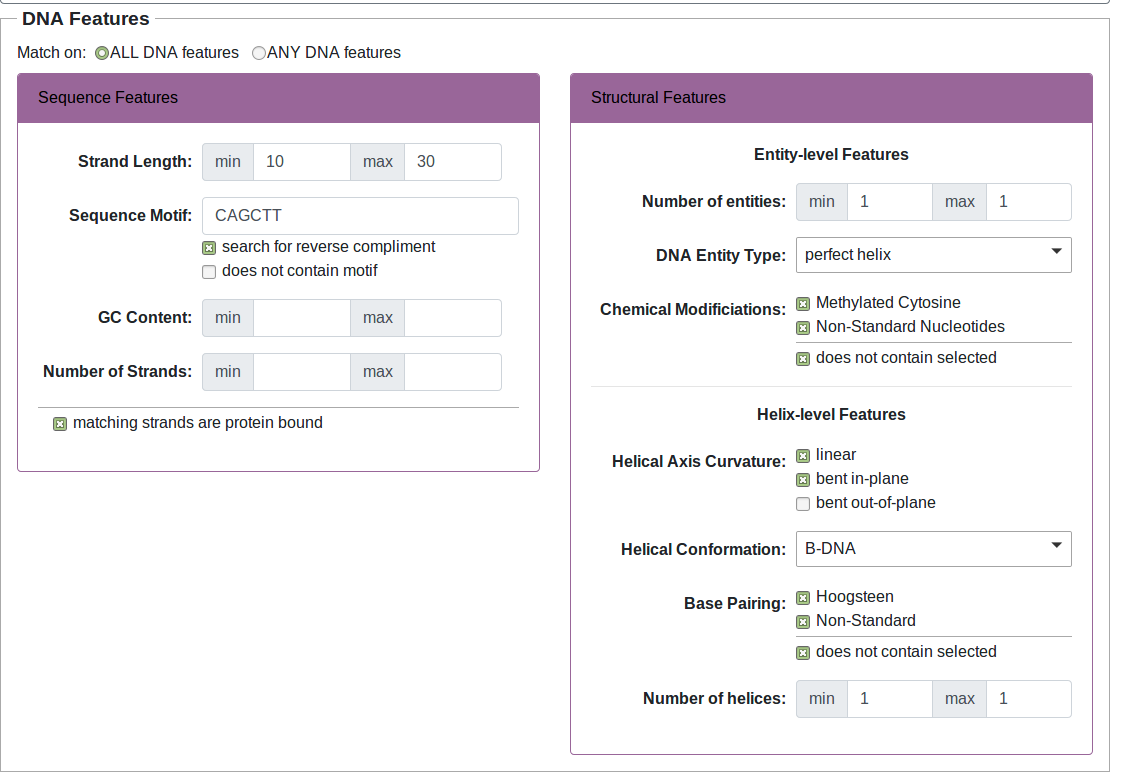
- Search thousands of DNA-protein complex structures based on features of the DNA, protein or DNA-protein interactions
- Visualize data using customizable, interactive visualizations. These can be exported for use in publications, or used as a data exploration tool
- Upload your own structures obtained from experiment or simulation and process them with our pipeline to generate the same data and visualizations which are available for any structure in our database.
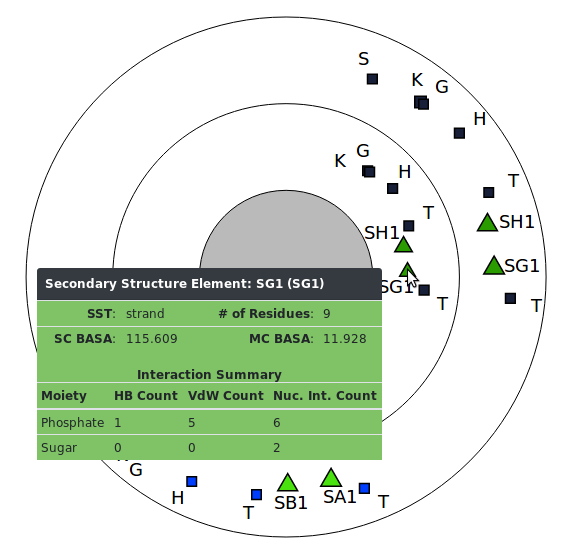
The data you retrieve can be downloaded directly for your use, or can be visualized with our built-in visualization tools which offer both customization and interactivity. Use the navigation bar at the top of the page to explore different areas of the site. Full documentation is available at the documentation page.
If you use DNAproDB in your work, please cite us: Jared M. Sagendorf, Helen M. Berman, Remo Rohs; Nucleic Acids Res 2017, 45, W89-W97. doi: 10.1093/nar/gkx272 and Jared M. Sagendorf, Nicholas Markarian, Helen M. Berman, Remo Rohs; Nucleic Acids Res 2019, 48, D277-D287. doi: 10.1093/nar/gkz889
Latest PDB Data Retrieval:
Latest DNAproDB Update:
DNAproDB provides data on structures retrieved from the PDB which contain at least one protein chain bound to at least one DNA strand of length two nucleotides or more. Very large structures (> 3000 components, i.e. residues or nucleotides) are currently not included.